Enriching Metabolic PATHwaY models with evidence from the literature (EMPATHY)
Background
Metabolic network reconstructions, along with their analysis using constraint-based approaches, are becoming increasingly important across a range of application areas, from human health, drug target discovery and drug repositioning, through to metabolic engineering and synthetic biology. The construction of genome-scale metabolic models has to date proven to be both expensive and time-consuming, often requiring further refinement as additional biochemical knowledge is deposited into data repositories and, more importantly, the literature. The main limitation in the development of such models is the extraction of this biochemical knowledge from the literature, traditionally carried out by a community of researchers as part of a "jamboree" approach (i.e., face-to-face meetings lasting two or three days), which although successful, is unsustainable given the increasing number of models requiring refinement and the increasing volume of literature being published.
Model construction requires complex evidence from the literature, since individual findings must be considered within their biological context (organism, cell type) to ensure that they are interpreted correctly. Text mining has been used to extract new knowledge hidden in text, enabling scientists to collect, interpret and discover knowledge needed for research, in an efficient and systematic manner. Integration of accurate text mining methods into a metabolic network reconstruction platform will help in establishing literature-based evidence (as in our previous work on PathText) and will facilitate the curation of reconstructions, thus dramatically increasing their coverage.
Project Objectives
The EMPATHY project aims to support metobolic pathway model curation through the integration of text mining methodologies into a pathway reconstruction platform. Specifically, we set out to accomplish the following:
- creation of a web-based platform that will allow users to develop their reconstructions using a graphical, user-interactive interface
- development of advanced text mining (TM) methods for extracting information on metabolic reactions from literature
- integration of TM methods into the reconstruction platform to facilitate the automatic provision of literature-based evidence and revision suggestions to the user
- development of an active learning-like mechanism that iteratively captures a user's feedback on text-mined evidence/suggestions and recalibrates the underlying tools in order to produce improved results
Framework
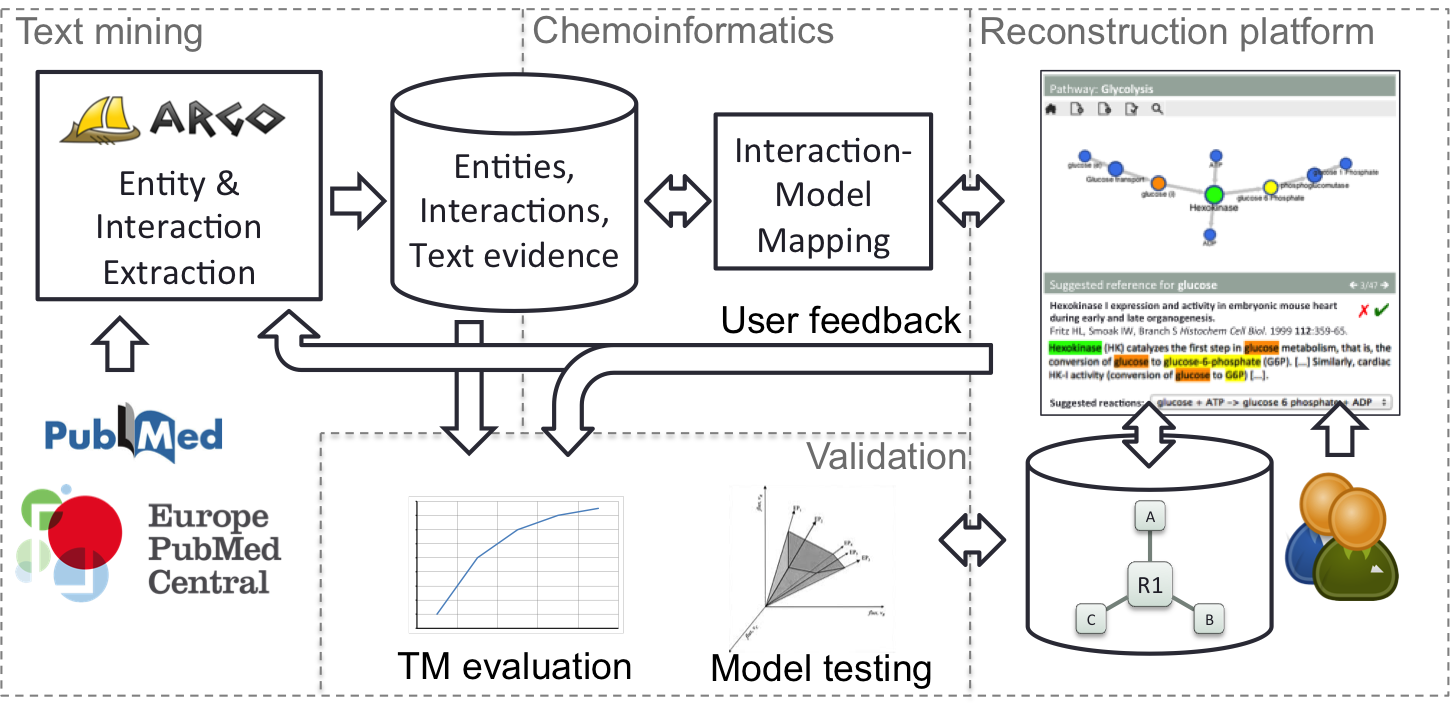
The figure above depicts EMPATHY's overall architecture and shows four inter-related components:
- Reconstruction platform. We shall develop a web application that will serve as a metabolic network reconstruction platform. Its backend will be populated with existing models from sources such as Recon 2, a large-scale, community-developed, "Google Map" of human metabolism. Users are provided with a user-interactive graphical interface for creating, managing and revising reconstructions.
- Text mining. Information on metabolic reactions will be mined from scientific literature using methods for extracting named entities (e.g., metabolites, enzymes) and interactions between them. These methods will be developed by means of our web-based text mining workbench, Argo, which integrates various natural language processing and machine learning tools into processing workflows.
- Chemoinformatics. Text-mined information will be analysed by chemoinformatics-based methods in order to map them with a reconstruction's relevant sections. In this way, literature-based evidence or suggestions are automatically presented to the user, thus enabling the more efficient curation of reconstructions..
- Validation. Our text mining methods will be evaluated using standard measures of performance (e.g., precision, recall). The overall framework will be tested by further developing models of three key organisms (human, yeast, E. coli), which will be evaluated against both automatically generated models and equivalent, hand-curated models.
News
30th November - 1st December 2017
Prof. Sophia Ananiadou will give a talk entitled Machine reading for cancer biology at the Global Pharma R&D Informatics Congress in Lisbon, Portugal.
May 2016 - The EMPATHY project is mentioned in a new article about text mining and the work of NaCTeM, published in Pharma Technology Focus, a bi-monthly magazine that brings together the latest insights and innovations from across the pharaceutical industry.
Publications
Swainston, N., Batista-Navarro, R., Carbonel, P., Dobson, P., Dunstan, M., Jervis, A., Vinaixa, M., Williams, A., Ananiadou, S., Faulon, J. L., Mendes, P., Kell, D. B., Scrutton, N. and Breitling, R. (2017). biochem4j: integrated and extensible biochemical knowledge through graph databases. PLOS ONE, 12(7), e0179130
Zerva, C., Batista-Navarro, R., Day, P. J. R. and Ananiadou, S. (2017). Using uncertainty to link and rank evidence from biomedical literature for model curation. Bioinformatics
Batista-Navarro, R. and Ananiadou, S. (2017). A text mining-based approach to graph database curation in support of metabolic pathway model reconstruction. Proceedings of Biocuration 2017
Przybyla, P., Shardlow, M., Aubin, S., Bossy, R., Eckart de Castilho, R., Piperidis, S., McNaught, J. and Ananiadou, S. (2016). Text Mining Resources for the Life Sciences. Database: The Journal of Biological Databases and Curation: baw145
References
Miwa, M., Ohta, T., Rak, R., Rowley, A., Kell, D. B., Pyysalo, S. and Ananiadou, S. (2013). A method for integrating and ranking the evidence for biochemical pathways by mining reactions from text. Bioinformatics, 29(13), i44-i52
Rak, R., Rowley, A., Black, W.J. and Ananiadou, S. (2012). Argo: an integrative, interactive, text mining-based workbench supporting curation. Database: The Journal of Biological Databases and Curation, 2012
Thiele, I., Swainston, N., Fleming, R. M. T., Hoppe, A., Sahoo, S.,Aurich, M. K., Haraldsdottir, H., et al. (2013). A community-driven global reconstruction of human metabolism.. Nature Biotechnology 31, 419-425
Project Team
Principal Investigator: Prof. Sophia Ananiadou (NaCTeM, School of Computer Science, University of Manchester)
Co-Investigators:
Prof. Douglas B. Kell (School of Chemistry, University of Manchester)
Dr. Neil Swainston (School of Computer Science, University of Manchester)
Researchers:
Dr. Paul Dobson (School of Computer Science, University of Manchester)
Mr. Sunil Sahu (NaCTeM, School of Computer Science, University of Manchester)
Funding
This project, which runs from April 2015 until April 2018, is being funded by the BBSRC (Grant No. BB/M006891/1)
Featured News
- AI for Research: How Can AI Disrupt the Research Process?
- ELLIS Workshop on Misinformation Detection - Presentation slides now available
- 1st Workshop on Misinformation Detection in the Era of LLMs (MisD)- 23rd June 2025
- Prof. Sophia Ananiadou accepted as an ELLIS fellow
- Invited talk at the 15th Marbach Castle Drug-Drug Interaction Workshop
- BioNLP 2025 and Shared Tasks accepted for co-location at ACL 2025
- Prof. Junichi Tsujii honoured as Person of Cultural Merit in Japan
- Participation in panel at Cyber Greece 2024 Conference, Athens
- New Named Entity Corpus for Occupational Substance Exposure Assessment
Other News & Events
- CL4Health @ NAACL 2025 - Extended submission deadline - 04/02/2025
- Shared Task on Financial Misinformation Detection at FinNLP-FNP-LLMFinLegal
- FinNLP-FNP-LLMFinLegal @ COLING-2025 - Call for papers
- Keynote talk at Manchester Law and Technology Conference
- Keynote talk at ACM Summer School on Data Science, Athens








