Big Mechanism
Background
Big mechanisms are large, explanatory models of complicated systems in which interactions have important causal effects. A greater understanding of such mechanisms is of particular importance for research into cancer biology, since it is not fully understood how to stop cancer cells from growing faster than normal cells. Given that most cancer drugs are highly toxic, it is important to find drug combinations that are tailored to individual patients and their cancer's genotypes, and which facilitate intervention at multiple points along a cancer pathway.
Project aims
This project, one of a number funded by DARPA as part of their Big Mechanism programme, aims to address the issues introduced above by automating the process of intelligent, optimised drug discovery in cancer research. This will be achieved through the application of a number of different techniques:
- Text Mining (TM) techniques will be developed to locate, extract, interpret and assimilate relevant nuggets of information which are fragmented and scattered throughout the vast volumes of potentially relevant literature, in order to build up a detailed background knowledge about causal models of cancer mechanisms
- Claims about cancer extracted by the TM analysis will be used to populate custom-designed ontologies to enable computational modelling and integration of information relating to cancer mechanisms and pathways
- Probabilistic reasoning over these models will facilitate automated hypothesis generation to strategically extend the knowledge
- A 'Robot Scientist' will perform experiments to test hypotheses and feed results back into the system
A combination of TM techniques can allow the recognition in literature of:
- entities, such as diseases, drugs and proteins
- causal relationships (or events) that hold between them, e.g., inhibition, activation, pathological processes, etc.
- information about the interpretation of bio-processes (e.g., negation, speculation, information provenance, etc). Such information will be important in assigning probabilities to bio-processes and detecting potential conflicts in the literature
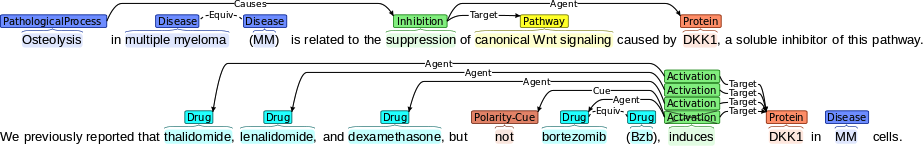
Example showing the types of entities, events and interpretational information which are intended to be recognised by TM techniques
Related Work
The TM techniques developed by NaCTeM will build upon a sucessful body of previous work, including the following:- The EventMine system (Miwa et al., 2012a, Miwa et al., 2012b), which automatically identifies biomedical events. In the context of the BioNLP shared tasks (2009, 2011, 2013), EventMine has been shown to obtain state-of-the-art performance in several different domains, including cancer genetics (Pyysalo et al., 2013; Miwa and Ananiadou, 2013)
- A collaborative project with AstraZenca, whose aim was to help enhance decision-making in drug discovery. A major focus was on the development of annotated corpora to facilitate the extraction of information relating to angiogenesis, i.e., the development of new blood vessels from existing ones, which is an area of high interest in cancer research.
- A corpus and system to facilitate the automatic extraction of terms and events relating to angiogenesis (Wang et al., 2011)
- The Multi-Level Event Extraction (MLEE) corpus (Pyysalo et al., 2012), consisting of abstracts of publications on angiogenesis, manually anotated with over 8,000 entities with fine-grained types and over 6,000 events.
- A revised version of FACTA+ (Tsuruoka et al., 2011), NaCTeM's system for finding associations between biomedical entities, which was updated to allow relationships between angiogenesis-associated genes and other biomedical entities to be identified.
- The PathText 2 system (Miwa et al., 2013), which associates pathway model reactions with relevant publications.
- The BioCause corpus (Mihaila et al., 2013), which aims to facilitate the development of systems that can automatically detect causal relationships in biomedical text
- The meta-knowledge corpus (Thompson et al., 2011), whose aim is to allow systems to recognise information pertaining to the interpretation of biomedical events (e.g., negation, speculation and knowledge source)
- New! - EUPMC Evidence Finder for Anantomical entities with meta-knowledge – A system that facilitates searching for facts relating to anatomical entities in full text articles. Facts can be filtered according to various meta-knowledge aspects (e.g., negation, certainly level, novelty)
- Argo (Rak et al., 2012), a collaborative, interoperable workbench, which allows TM processing pipelines to be built and evaluated with mininal effort
- The U-Compare library (Kano et al., 2011) of interoperable TM processing tools, including ones tailored for processing biomedical text.
News
30th November - 1st December 2017
Prof. Sophia Ananiadou will give a talk entitled Machine reading for cancer biology at the Global Pharma R&D Informatics Congress in Lisbon, Portugal.
26th - 29th March 2017
A workshop relating to work being carried on the Big Mechanism project will form part of the scientific programme at the 10th International Biocuration conference conference, to be held at Stanford University
Workshop Title: Reading, Assembling and Reasoning for Biocuration
Organizers: Sophia Ananiadou, Riza Batista-Navarro, Paul Cohen, Diana Chung, Emek Demir, Lynette Hirschman, Parag Mallik
Summary: We will focus on recent advances in the development of integrated systems to capture "Big Mechanisms" for biological systems, including machine reading of journal articles, (semi-)automated assembly of signaling pathway models, and machine-aided analysis of these models for tasks such as drug repurposing and explaining drugs' effects. This workshop will consist of invited speakers and contributed talks and/or panel discussions from experts in biocuration, machine reading, and biological modeling.
20th May 2016
The Big Mechanism project is mentioned in a new article about text mining and the work of NaCTeM, published in Pharma Technology Focus, a bi-monthly magazine that brings together the latest insights and innovations from across the pharaceutical industry.
5th April 2016
Prof. Sophia Ananiadou will give a seminar entitled Text Mining tools and infrastructure for biomedical applications - cancer biology, history of medicine, monitoring biodiversity at the CERTH Conference Centre Vergina, Greece.
Project Publications
Shardlow, M., Batista-Navarro, R., Thompson, P., Nawaz, R., McNaught, J. and Ananiadou, S. (2018). Identification of research hypotheses and new knowledge from scientific literature. BMC medical informatics and decision making, 18(1), 46.
Przybyła, P., Soto, A. and Ananiadou, S. (2017). Identifying Personalised Treatments and Clinical Trials for Precision Medicine using Semantic Search with Thalia. In Proceedings of the Twenty-Fifth Text REtrieval Conference (TREC 2017).
Soto, A. J., Zerva, C., Batista-Navarro, R. and Ananiadou, S. (2017). LitPathExplorer: a confidence-based visual text analytics tool for exploring literature-enriched pathway models. Bioinformatics, 34(8), 1389-1397.
Zerva, C., Batista-Navarro, R., Day, P., and Ananiadou, S. (2017). Using uncertainty to link and rank evidence from biomedical literature for model curation. Bioinformatics, 33(23), 3784-3792.
Alnazzawi, N., Thompson, P. and Ananiadou, S. (2016). Mapping Phenotypic Information in Heterogeneous Textual Sources to a Domain-Specific Terminological Resource. PLOS ONE, 11(9), e0162287.
Alnazzawi, N., Thompson, P., Batista-Navarro, R. T. B. and Ananiadou, S. (2015). Using text mining techniques to extract phenotypic information from the PhenoCHF corpus. BMC Medical Informatics and Decision Making, 15(Suppl. 2):S3
Batista-Navarro, R. T. B. and Ananiadou, S.(2015). Augmenting the Medical Subject Headings vocabulary with semantically rich variants to improve disease mention normalisation. In Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, Seville, Spain, pp. 311-316
Batista-Navarro, R. T. B., Carter, J. and Ananiadou, S. (2015). Development of bespoke machine learning and biocuration workflows in a BioC-supporting text mining workbench. In Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, Seville, Spain, pp. 51-56
Batista-Navarro, R. T. B., Carter, J. and Ananiadou, S. (2015). Semi-automatic curation of chronic obstructive pulmonary disease phenotypes using Argo. In Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, Seville, Spain, pp. 403-408
Fu, X., Batista-Navarro, R. T. B., Rak, R. and Ananiadou, S. (2015). Supporting the Annotation of Chronic Obstructive Pulmonary Disease (COPD) Phenotypes with Text Mining Workflows. Journal of Biomedical Semantics, 6:8 (Highly Accessed)
Xu, Y., Chen, L., Wei, J., Ananiadou, S., Fan, Y., Qian, Y., Chang, E. I-C. and Tsujii, J. (2015). Bilingual term alignment from comparable corpora in English discharge summary and Chinese discharge summary. BMC Bioinformatics 16:149.
Zerva, C. and Ananiadou, S. (2015). Event Extraction in pieces: Tackling the partial event identification problem on unseen corpora. In Proceedings of the 2015 Workshop on Biomedical Natural Language Processing (BioNLP 2015), pp. 31-41
Xu, Y., Hua, J., Ni, Z., Chen, Q., Fan, Y., Ananiadou, S., Chang, E. I-C. and Tsujii, J. (2014). Anatomical entity recognition with a hierarchical framework augmented by external resources PLOS ONE, 9(10), e108396
References
Kano, Y., Miwa, M., Cohen, K. B., Hunter, L., Ananiadou, S. and Tsujii, J. (2011). U-Compare: a modular NLP workflow construction and evaluation system. IBM Journal of Research and Development, 55(3), 11:1 - 11:10
Mihaila, C., Ohta, T., Pyysalo, S. and Ananiadou, S. (2013). BioCause: Annotating and Analysing Causality in the Biomedical Domain. BMC Bioinformatics, 14:2
Miwa, M., Thompson, P. and Ananiadou, S.. (2012a). Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. In: Bioinformatics, 28(13), 1759-1765
Miwa, M., Thompson, P., McNaught, J., Kell, D. B. and Ananiadou, S.. (2012b). Extracting semantically enriched events from biomedical literature. In: BMC Bioinformatics, 13, 108
Miwa, M. and Ananiadou, S. (2013). NaCTeM EventMine for BioNLP 2013 CG and PC tasks. Proceedings of BioNLP Shared Task 2013 Workshop, Sofia, Bulgaria, pp. 94-98.
Miwa, M., Ohta, T., Rak, R., Rowley, A., Kell, D. B., Pyysalo, S. and Ananiadou, S. (2013). A method for integrating and ranking the evidence for biochemical pathways by mining reactions from text. Bioinformatics, 29(13), i44-i52
Pyysalo, S., Ohta, T., Miwa, M., Cho, H. -C., Tsujii, J. and Ananiadou, S. (2012). Event extraction across multiple levels of biological organization. In: Bioinformatics, 28(18), i575-i581
Pyysalo, S., Ohta, T. and Ananiadou, S. (2013). Overview of the Cancer Genetics (CG) task of BioNLP Shared Task 2013. Proceedings of the BioNLP Shared Task 2013 Workshop, Sofia, Bulgaria, pp. 58-66, Association for Computational Linguistics
Rak, R., Rowley, A., Black, W.J. and Ananiadou, S.. (2012). Argo: an integrative, interactive, text mining-based workbench supporting curation. Database: The Journal of Biological Databases and Curation, 2012
Thompson, P., Nawaz, R., McNaught, J. and Ananiadou, S. (2011). Enriching a biomedical event corpus with meta-knowledge annotation. BMC Bioinformatics, 12, 393
Tsuruoka, Y., Miwa, M., Hamamoto, K., Tsujii, J. and Ananiadou, S. (2011). Discovering and visualizing indirect associations between biomedical concepts. In: Bioinformatics, 27(13), i111-i119
Wang, X., McKendrick, I., Barrett, I., Dix, I., French, T., Tsujii, J. and Ananiadou, S.. (2011). Automatic Extraction of Angiogenesis Bio-Process from Text. In: Bioinformatics, 27(19), 2730-2737
Project team
Principal Investigator: Prof. Andrey Rzhetsky, Section of Genetic Medicine,University of Chicago, USACo-Investigators:
Prof. Sophia Ananiadou (NaCTeM)
Prof. Ross King (School of Computer Science, University of Manchester)
Dr. Larisa Soldatova (School of Information Systems, Computing and Mathematics, Brunel University, London)
Prof. Robert Stevens (School of Computer Science, University of Manchester)
Prof. Jun'ichi Tsujii (Microsoft Research Asia, Beijing, China)
Dr. Hoifung Poon (Mircrosoft Research, Redmond, WA, USA)
Featured News
- 1st Workshop on Misinformation Detection in the Era of LLMs - Presentation slides now available
- Prof. Ananiadou appointed Deputy Director of the Christabel Pankhurst Institute
- ELLIS Workshop on Misinformation Detection - Presentation slides now available
- Prof. Sophia Ananiadou accepted as an ELLIS fellow
- BioNLP 2025 and Shared Tasks accepted for co-location at ACL 2025
- Prof. Junichi Tsujii honoured as Person of Cultural Merit in Japan
Other News & Events
- AI for Research: How Can AI Disrupt the Research Process?
- CL4Health @ NAACL 2025 - Extended submission deadline - 04/02/2025
- Invited talk at the 15th Marbach Castle Drug-Drug Interaction Workshop
- Participation in panel at Cyber Greece 2024 Conference, Athens
- Shared Task on Financial Misinformation Detection at FinNLP-FNP-LLMFinLegal








